Post processing
- biapy.data.post_processing.post_processing.boundary_refinement_watershed(X, Y_pred, erode=True, save_marks_dir=None)[source]
Apply watershed to the given predictions with the goal of refine the boundaries of the artifacts.
Based on https://docs.opencv.org/master/d3/db4/tutorial_py_watershed.html.
- Parameters:
X (4D Numpy array) – Original data to guide the watershed. E.g.
(img_number, y, x, channels).Y_pred (4D Numpy array) – Predicted data to refine the boundaries. E.g.
(img_number, y, x, channels).erode (bool, optional) – To extract the sure foreground eroding the artifacts instead of doing with distanceTransform.
save_marks_dir (str, optional) – Directory to save the markers used to make the watershed. Useful for debugging.
- Returns:
Array – Refined boundaries of the predictions. E.g.
(img_number, y, x, channels).- Return type:
4D Numpy array
Examples
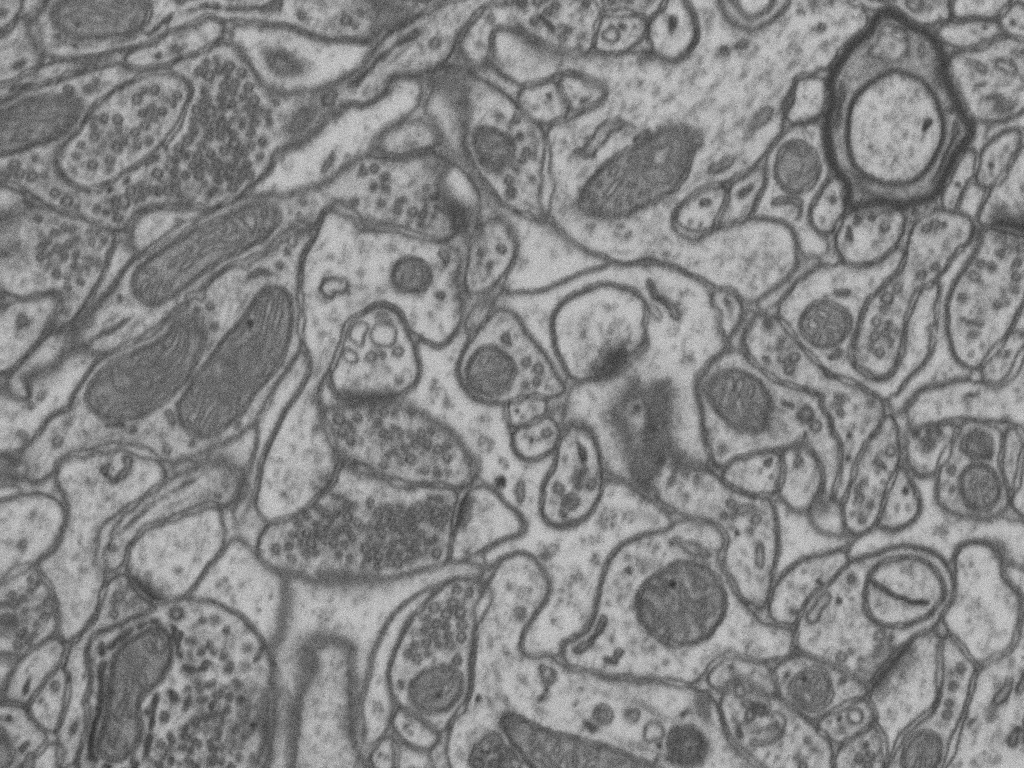
Original image

Ground truth

Predicted image

Watershed ouput
The marks used to guide the watershed is this example are these:
- biapy.data.post_processing.post_processing.boundary_refinement_watershed2(X, Y_pred, save_marks_dir=None)[source]
Apply watershed to the given predictions with the goal of refine the boundaries of the artifacts. This function was implemented using scikit instead of opencv as
post_processing.boundary_refinement_watershed().Based on https://scikit-image.org/docs/dev/auto_examples/segmentation/plot_watershed.html.
- Parameters:
X (4D Numpy array) – Original data to guide the watershed. E.g.
(img_number, y, x, channels).Y_pred (4D Numpy array) – Predicted data to refine the boundaries. E.g.
(img_number, y, x, channels).save_marks_dir (str, optional) – Directory to save the markers used to make the watershed. Useful for debugging.
- Returns:
Array – Refined boundaries of the predictions. E.g.
(img_number, y, x, channels).- Return type:
4D Numpy array
- biapy.data.post_processing.post_processing.watershed_by_channels(data, channels, ths={}, remove_before=False, thres_small_before=10, seed_morph_sequence=[], seed_morph_radius=[], erode_and_dilate_foreground=False, fore_erosion_radius=5, fore_dilation_radius=5, rmv_close_points=False, remove_close_points_radius=-1, resolution=[1, 1, 1], watershed_by_2d_slices=False, save_dir=None)[source]
Convert binary foreground probability maps and instance contours to instance masks via watershed segmentation algorithm.
Implementation based on PyTorch Connectomics’ process.py.
- Parameters:
data (4D Numpy array) – Binary foreground labels and contours data to apply watershed into. E.g.
(397, 1450, 2000, 2).channels (str) – Channel type used. Possible options:
BC,BCM,BCD,BCDv2,Dv2andBDv2.ths (float, optional) – Thresholds to be used on each channel.
TH_BINARY_MASKused in the semantic mask to create watershed seeds;TH_CONTOURused in the contours to create watershed seeds;TH_FOREGROUNDused in the semantic mask to create the foreground mask;TH_POINTSused in the point mask to create watershed seeds;TH_DISTANCEused in the distances to create watershed seeds.remove_before (bool, optional) – To remove objects before watershed.
thres_small_before (int, optional) – Theshold to remove small objects created by the watershed.
seed_morph_sequence (List of str, optional) – List of strings to determine the morphological filters to apply to instance seeds. They will be done in that order. E.g.
['dilate','erode'].seed_morph_radius (List of ints, optional) – List of ints to determine the radius of the erosion or dilation for instance seeds.
erode_and_dilate_foreground (bool, optional) – To erode and dilate the foreground mask before using marker controlled watershed. The idea is to remove the small holes that may be produced so the instances grow without them.
fore_erosion_radius (int, optional) – Radius to erode the foreground mask.
fore_dilation_radius (int, optional) – Radius to dilate the foreground mask.
rmv_close_points (bool, optional) – To remove close points to each other. Used in ‘BP’ channel configuration.
remove_close_points_radius (float, optional) – Radius from each point to decide what points to keep. Used in ‘BP’ channel configuration. E.g.
10.0.resolution (ndarray of floats) – Resolution of the data, in
(z,y,x)to calibrate coordinates. E.g.[30,8,8].watershed_by_2d_slices (bool, optional) – Whether to apply or not the watershed to create instances slice by slice in a 3D problem. This can solve instances invading others if the objects in Z axis overlap too much.
save_dir (str, optional) – Directory to save watershed output into.
- biapy.data.post_processing.post_processing.calculate_zy_filtering(data, mf_size=5)[source]
Applies a median filtering in the z and y axes of the provided data.
- Parameters:
data (4D Numpy array) – Data to apply the filter to. E.g.
(num_of_images, y, x, channels).mf_size (int, optional) – Size of the median filter. Must be an odd number.
- Returns:
Array – Filtered data. E.g.
(num_of_images, y, x, channels).- Return type:
4D Numpy array
- biapy.data.post_processing.post_processing.calculate_z_filtering(data, mf_size=5)[source]
Applies a median filtering in the z dimension of the provided data.
- Parameters:
data (4D Numpy array) – Data to apply the filter to. E.g.
(num_of_images, y, x, channels).mf_size (int, optional) – Size of the median filter. Must be an odd number.
- Returns:
Array – Filtered data. E.g.
(num_of_images, y, x, channels).- Return type:
4D Numpy array
- biapy.data.post_processing.post_processing.ensemble8_2d_predictions(o_img, pred_func, axis_order_back, axis_order, device, batch_size_value=1)[source]
Outputs the mean prediction of a given image generating its 8 possible rotations and flips.
- Parameters:
o_img (3D Numpy array) – Input image. E.g.
(y, x, channels).pred_func (function) – Function to make predictions.
axis_order_back (tuple) – Axis order to convert from tensor to numpy. E.g.
(0,3,1,2).axis_order (tuple) – Axis order to convert from numpy to tensor. E.g.
(0,3,1,2).device (Torch device) – Device used.
batch_size_value (int, optional) – Batch size value.
- Returns:
out – Output image ensembled. E.g.
(y, x, channels).- Return type:
3D Numpy array
Examples
# EXAMPLE 1 # Apply ensemble to each image of X_test X_test = np.ones((165, 768, 1024, 1)) out_X_test = np.zeros(X_test.shape, dtype=(np.float32)) for i in tqdm(range(X_test.shape[0])): pred_ensembled = ensemble8_2d_predictions(X_test[i], pred_func=(lambda img_batch_subdiv: model(img_batch_subdiv)), n_classes=n_classes) out_X_test[i] = pred_ensembled
- biapy.data.post_processing.post_processing.ensemble16_3d_predictions(vol, pred_func, axis_order_back, axis_order, device, batch_size_value=1)[source]
Outputs the mean prediction of a given image generating its 16 possible rotations and flips.
- Parameters:
o_img (4D Numpy array) – Input image. E.g.
(z, y, x, channels).pred_func (function) – Function to make predictions.
axis_order_back (tuple) – Axis order to convert from tensor to numpy. E.g.
(0,3,1,2,4).axis_order (tuple) – Axis order to convert from numpy to tensor. E.g.
(0,3,1,2,4).device (Torch device) – Device used.
batch_size_value (int, optional) – Batch size value.
- Returns:
out – Output image ensembled. E.g.
(z, y, x, channels).- Return type:
4D Numpy array
Examples
# EXAMPLE 1 # Apply ensemble to each image of X_test X_test = np.ones((10, 165, 768, 1024, 1)) out_X_test = np.zeros(X_test.shape, dtype=(np.float32)) for i in tqdm(range(X_test.shape[0])): pred_ensembled = ensemble8_2d_predictions(X_test[i], pred_func=(lambda img_batch_subdiv: model(img_batch_subdiv))) out_X_test[i] = pred_ensembled
- biapy.data.post_processing.post_processing.create_th_plot(ths, y_list, th_name='TH_BINARY_MASK', chart_dir=None, per_sample=True, ideal_value=None)[source]
Create plots for threshold value calculation.
- Parameters:
ths (List of floats) – List of thresholds. It will be the
xaxis.y_list (List of ints/floats) – Values of
yaxis.th_name (str, optional) – Name of the threshold.
chart_dir (str, optional) – Path where the charts are stored.
per_sample (bool, optional) – Create the plot per list in
y_list.ideal_value (int/float, optional) – Value that should be the ideal optimum. It is going to be marked with a red line in the chart.
- biapy.data.post_processing.post_processing.voronoi_on_mask(data, mask, th=0, verbose=False)[source]
Apply Voronoi to the voxels not labeled yet marked by the mask. It is done using distances from the un-labeled voxels to the cell perimeters.
- Parameters:
data (2D/3D Numpy array) – Data to apply Voronoi.
(y, x)for 2D or(z, y, x)for 3D. E.g.(397, 1450, 2000)for 3D.mask (3D/4D Numpy array) – Data mask to determine which points need to be proccessed.
(z, y, x, channels)e.g.(397, 1450, 2000, 3).th (float, optional) – Threshold used to binarize the input. If th=0, otsu threshold is used.
thres_small (int, optional) – Theshold to remove small objects created by the watershed.
verbose (bool, optional) – To print saving information.
- Returns:
data – Image with Voronoi applied.
(num_of_images, z, y, x)e.g.(1, 397, 1450, 2000)- Return type:
4D Numpy array
- biapy.data.post_processing.post_processing.remove_close_points(points, radius, resolution, classes=None, ndim=3, return_drops=False)[source]
Remove all points from
point_listthat are at aradiusor less distance from each other.- Parameters:
points (ndarray of floats) – List of 3D points. E.g.
((0,0,0), (1,1,1).radius (float) – Radius from each point to decide what points to keep. E.g.
10.0.resolution (ndarray of floats) – Resolution of the data, in
(z,y,x)to calibrate coordinates. E.g.[30,8,8].ndim (int, optional) – Number of dimension of the data.
return_drops (bool, optional) – Whether to return or not a list containing the positions of the points removed.
- Returns:
new_point_list – New list of points after removing those at a distance of
radiusor less from each other.- Return type:
List of floats
- biapy.data.post_processing.post_processing.detection_watershed(seeds, coords, data_filename, first_dilation, nclasses=1, ndim=3, donuts_classes=[-1], donuts_patch=[13, 120, 120], donuts_nucleus_diameter=30, save_dir=None)[source]
Grow given detection seeds.
- Parameters:
seeds (4D Numpy array) – Binary foreground labels and contours data to apply watershed into. E.g.
(397, 1450, 2000, 2).coords (List of 3 ints) – Coordinates of all detected points.
data_filename (str) – Path to load the image paired with seeds.
first_dilation (str) – Each class seed’s dilation before watershed.
nclasses (int, optional) – Number of classes.
ndim (int, optional) – Number of dimensions. E.g. for 2D set it to
2and for 3D to3.donuts_classes (List of ints, optional) – Classes to check a donuts type cell. Set to
-1to disable it.donuts_patch (List of ints, optional) – Patch to analize donuts cells. Give a shape that covers all sizes of this type of cells.
donuts_nucleus_diameter (int, optional) – Aproximate nucleus diameter for donuts type cells.
save_dir (str, optional) – Directory to save watershed output into.
- Returns:
segm – Image with Voronoi applied.
(num_of_images, z, y, x)e.g.(1, 397, 1450, 2000)- Return type:
4D Numpy array
- biapy.data.post_processing.post_processing.measure_morphological_props_and_filter(img, resolution, filter_instances=False, properties=[[]], prop_values=[[]], comp_signs=[[]])[source]
Measures the properties of input image’s instances. It calculates each instance id, number of pixels, area/volume (2D/3D respec. and taking into account the resolution), diameter, perimeter/surface_area (2D/3D respec.), circularity/sphericity (2D/3D respec.) and elongation properties. All instances that satisfy the conditions composed by
properties,prop_valuesandcomp_signsvariables will be removed fromimg. Apart from returning all properties this function will return also a list identifying those instances that satisfy and not satify the conditions. Those removed will be marked as ‘Removed’ whereas the rest are ‘Correct’. Some of the properties follow the formulas used in MorphoLibJ library for Fiji.- Parameters:
img (2D/3D Numpy array) – Image with instances. E.g.
(1450, 2000)for 2D and(397, 1450, 2000)for 3D.resolution (str) – Path to load the image paired with seeds.
filter_instances (bool, optional) – Whether to do instance filtering or not.
properties (List of lists of str, optional) – List of lists of properties to remove the instances. Options available:
['circularity', 'npixels', 'area', 'diameter', 'elongation', 'sphericity', 'perimeter']. E.g.[['size'], ['circularity', 'npixels']].prop_values (List of lists of floats/ints, optional) – List of lists of values for each property. E.g.
[[70], [0.7, 2000]].comp_signs (List of list of str, optional) – List of lists of signs to compose the conditions, together
propertiesprop_values, that the instances must satify to be removed from the inputimg. E.g.[['le'], ['lt', 'ge']].
- Returns:
img (2D/3D Numpy array) – Input image without the instances that do not satisfy the circularity constraint. Image with instances. E.g.
(1450, 2000)for 2D and(397, 1450, 2000)for 3D.d_result (dict) – Results of the morphological measurements. All the information of the non-filtered instances (if declared to do so) are listed. It contains:
- labelsArray of ints
Instance label list.
- centersArray of ints
Coordinates of the centers of each instance.
- npixelsArray of ints
Number of pixels of each instance.
- areasArray of ints
Area/volume (2D/3D) of each instance based on the given resolution.
- circularitiesArray of ints
Circularity/sphericity (2D/3D) of each instance. In 2D,
circularityof an instance is defined as the ratio of area over the square of the perimeter, normalized such that the value for a disk equals one:(4 * PI * area) / (perimeter^2). While values of circularity range theoretically within the interval [0;1], the measurements errors of the perimeter may produce circularity values above 1 (Lehmann et al.). In 3D,sphericityis is the ratio of the squared volume over the cube of the surface area, normalized such that the value for a ball equals one:(36 * PI)*((volume^2)/(perimeter^3)).- diametersArray of ints
Diameter of each instance obtained from the bounding box.
- elongationsArray of ints
Elongation of each instance. It is the inverse of the circularity. The values of elongation range from
1for round particles and increase for elongated particles. Calculated as:(perimeter^2)/(4 * PI * area). Only measurable for 2D images.- perimeterArray of ints
In 2D, approximates the contour as a line through the centers of border pixels using a 4-connectivity. In 3D, it is the surface area computed using Lewiner et al. algorithm using marching_cubes and mesh_surface_area functions of scikit-image.
- commentList of str
List containing ‘Correct’ string when the instance surpass the circularity threshold and ‘Removed’ otherwise.
- conditionsList of str
List of conditions that each instance has satisfy or not.
- biapy.data.post_processing.post_processing.find_neighbors(img, label, neighbors=1)[source]
Find neighbors of a label in a given image.
- Parameters:
img (2D/3D Numpy array) – Image with instances. E.g.
(1450, 2000)for 2D and(397, 1450, 2000)for 3D.label (int) – Label to find the neighbors of.
neighbors (int, optional) – Number of neighbors in each axis to explore.
- Returns:
neighbors – Neighbors instance ids of the given label.
- Return type:
list of ints
- biapy.data.post_processing.post_processing.repare_large_blobs(img, size_th=10000)[source]
Try to repare large instances by merging neighbors ones with it and by removing possible central holes.
- Parameters:
img (2D/3D Numpy array) – Image with instances. E.g.
(1450, 2000)for 2D and(397, 1450, 2000)for 3D.size_th (int, optional) – Size that the instances need to be larger than to be analised.
- Returns:
img – Input image without the large instances repaired. E.g.
(1450, 2000)for 2D and(397, 1450, 2000)for 3D.- Return type:
2D/3D Numpy array
- biapy.data.post_processing.post_processing.apply_binary_mask(X, bin_mask_dir)[source]
Apply a binary mask to remove values outside it.
- Parameters:
X (3D/4D Numpy array) – Data to apply the mask. E.g.
(y, x, channels)for 2D or(z, y, x, channels)for 3D.bin_mask_dir (str, optional) – Directory where the binary mask are located.
- Returns:
X – Data with the mask applied. E.g.
(y, x, channels)for 2D or(z, y, x, channels)for 3D.- Return type:
3D/4D Numpy array
