(Paper) CartoCell, a high-throughput pipeline for accurate 3D image analysis
This tutorial describes how to train and infer using our custom ResU-Net 3D DNN in order to reproduce the results obtained in (Andrés-San Román, 2022). Given an initial training dataset of 21 segmented epithelial 3D cysts acquired after confocal microscopy, we follow the CartoCell pipeline (figure below) to high-throughput segment hundreds of cysts at low resolution automatically.
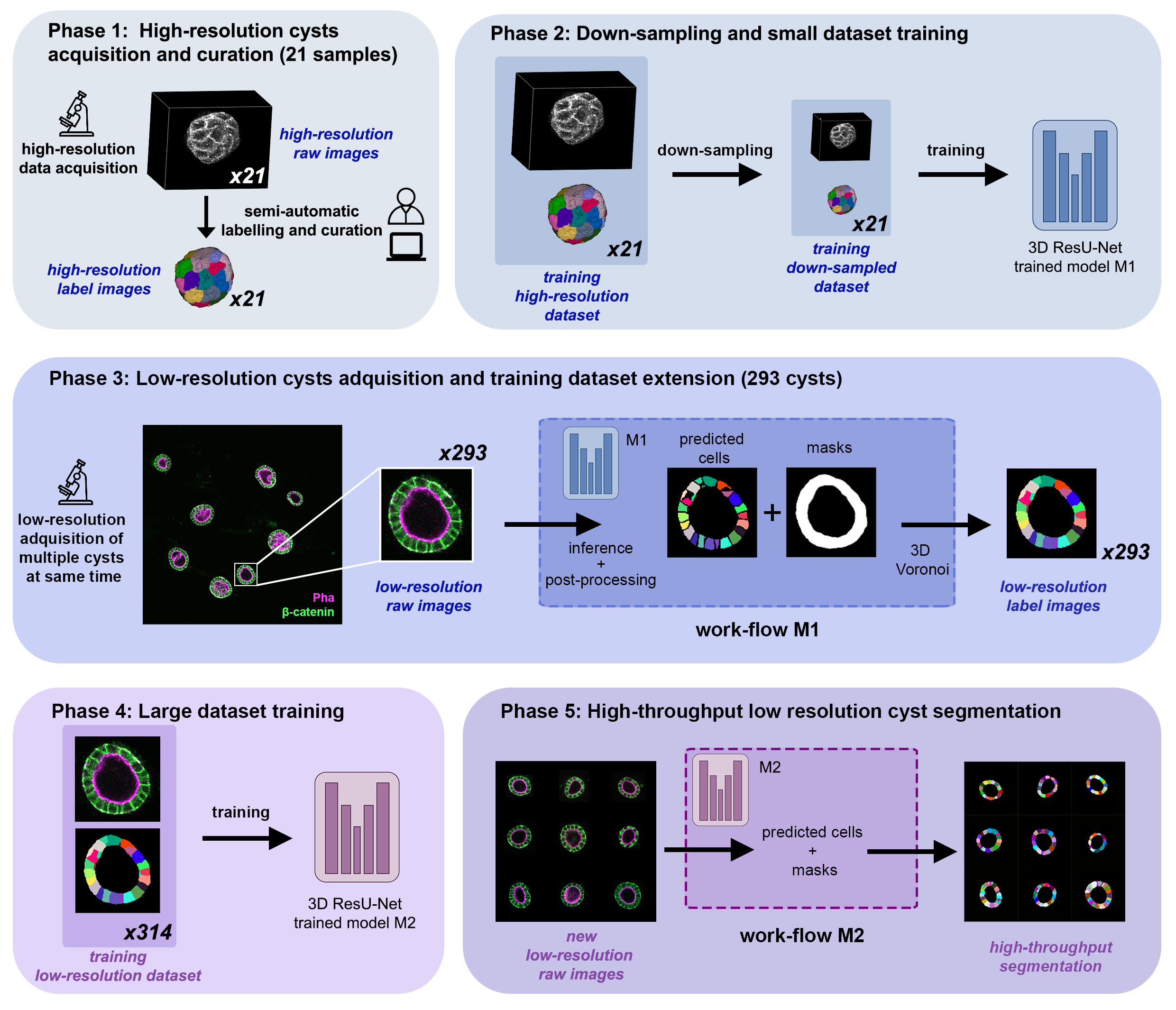
CartoCell pipeline for high-throughput epithelial cysts segmentation.
Cyst raw image |

Cyst label image |
Paper citation:
Andres-San Roman, Jesus A., et al. "CartoCell, a high-content pipeline for 3D image
analysis, unveils cell morphology patterns in epithelia." Cell Reports Methods
3.10 (2023).
CartoCell phases
A small dataset of 21 cysts, stained with cell outlines markers, was acquired at high-resolution in a confocal microscope. Next, the individual cell instances were segmented. The high-resolution images from Phase 1 provides the accurate and realistic set of data necessary for the following steps.
Both high-resolution raw and label images were down-sampled to create our initial training dataset. Specifically, image volumes were reduced to match the resolution of the images acquired in Phase 3. Using that dataset, a first DNN was trained. We will refer to this first model as model M1.
A large number of low-resolution stacks of multiple epithelial cysts was acquired. This was a key step to allow the high-throughput analysis of samples since it greatly reduces acquisition time. Here, we extracted the single-layer and single-lumen cysts by cropping them from the complete stack. This way, we obtained a set of 293 low-resolution images, composed of 84 cysts at 4 days, 113 cysts at 7 days and 96 cysts at 10 days. Next, we applied our trained model M1 to those images and post-processed their output to produce (i) a prediction of individual cell instances (obtained by marker-controlled watershed), and (ii) a prediction of the mask of the full cellular regions. At this stage, the output cell instances were generally not touching each other, which is a problem to study cell connectivity in epithelia. Therefore, we applied a 3D Voronoi algorithm to correctly mimic the epithelial packing. More specifically, each prediction of cell instances was used as a Voronoi seed, while the prediction of the mask of the cellular region defined the bounding territory that each cell could occupy. The result of this phase was a large dataset of low-resolution images and their corresponding accurate labels.
A new 3D ResU-Net model (model M2, from now on) was trained on the newly produced large dataset of low-resolution images and its paired label images. This was a crucial step, since the performance of deep learning models is highly dependent on the amount of training samples.
model M2 was applied to new low-resolution cysts and their output was post-processed as in Phase 3, thus achieving high-throughput segmentation of the desired cysts.
Data preparation
The data is accesible through Zenodo here. The data you need on each phase is:
train_M1 and validation to feed the initial model (model M1, Phase 2).
train_M2 to run Phase 3 – 5 of CartoCell pipeline.
test if you just want to run the inference using our pretrained model M2.
How to train your model
You have two options to train your model: via command line or using Google Colab.
You can reproduce the exact results of our manuscript via command line using cartocell_training.yaml configuration file.
In case you want to reproduce our model M1, Phase 2, you will need to modify the
DATA.TRAIN.PATHandDATA.TRAIN.GT_PATHwith the raw image and their corresponding labels, that is to say, with the paths of train_M1/x and train_M1/y respectively.In case you want to reproduce our model M2, Phase 4, you will need to modify the
DATA.TRAIN.PATHandDATA.TRAIN.GT_PATHas above but now using the paths of train_M2/x and train_M2/y.
For the validation data, for both model M1 and model M2, you will need to modify DATA.VAL.PATH and DATA.VAL.GT_PATH with the raw image and their corresponding labels, that is to say, with the paths of validation/x and validation/y respectively.
The next step is to open a terminal and run the code as follows:
# Configuration file
job_cfg_file=/home/user/cartocell_training.yaml
# Where the experiment output directory should be created
result_dir=/home/user/exp_results
# Just a name for the job
job_name=cartocell_training
# Number that should be increased when one need to run the same job multiple times (reproducibility)
job_counter=1
# Number of the GPU to run the job in (according to 'nvidia-smi' command)
gpu_number=0
# Move where BiaPy installation resides
git clone git@github.com:BiaPyX/BiaPy.git
cd BiaPy
git checkout 2bfa7508c36694e0977fdf2c828e3b424011e4b1
# Load the environment
conda activate BiaPy_env
python -u main.py \
--config $job_cfg_file \
--result_dir $result_dir \
--name $job_name \
--run_id $job_counter \
--gpu $gpu_number
Another alternative is to use a Google Colab . Noteworthy, Google Colab standard account do not allow you to run a long number of epochs due to time limitations. Because of this, we set
50 epochs to train and patience to 10 while the original configuration they are set to 1300 and 100 respectively. In this case you do not need to donwload any data, as the notebook will do it for you.
How to run the inference
You can reproduce the exact results of our model M2, Phase 5, of the manuscript via command line using cartocell_inference.yaml configuration file.
You will need to set DATA.TEST.PATH and DATA.TEST.GT_PATH with test/x and test/y data. You will need to download model_weights_cartocell.h5 file, which is the pretained model, and set its path in PATHS.CHECKPOINT_FILE.
The next step is to open a terminal and run the code as follows:
# Configuration file
job_cfg_file=/home/user/cartocell_inference.yaml
# Where the experiment output directory should be created
result_dir=/home/user/exp_results
# Just a name for the job
job_name=cartocell_inference
# Number that should be increased when one need to run the same job multiple times (reproducibility)
job_counter=1
# Number of the GPU to run the job in (according to 'nvidia-smi' command)
gpu_number=0
# Move where BiaPy installation resides (if you didn't in the previous steps)
git clone git@github.com:BiaPyX/BiaPy.git
cd BiaPy
git checkout 2bfa7508c36694e0977fdf2c828e3b424011e4b1
# Load the environment
conda activate BiaPy_env
python -u main.py \
--config $job_cfg_file \
--result_dir $result_dir \
--name $job_name \
--run_id $job_counter \
--gpu $gpu_number
Results
Following the example, the results should be placed in /home/user/exp_results/cartocell/results. You should find the following directory tree:
cartocell/
├── config_files/
| ├── cartocell_training.yaml
│ └── cartocell_inference.yaml
├── checkpoints
│ └── model_weights_cartocell_1.h5
└── results
└── cartocell_1
├── aug
│ └── .tif files
├── charts
│ ├── cartocell_1_jaccard_index.png
│ ├── cartocell_1_loss.png
│ └── model_plot_cartocell_1.png
├── per_image
│ └── .tif files
├── per_image_instances
│ └── .tif files
├── per_image_instances_voronoi
│ └── .tif files
└── watershed
├── seed_map.tif
├── foreground.tif
└── watershed.tif
config_files: directory where the .yaml filed used in the experiment is stored.cartocell_training.yaml: YAML configuration file used for training.cartocell_inference.yaml: YAML configuration file used for inference.
checkpoints: directory where model’s weights are stored.model_weights_cartocell_1.h5: model’s weights file.
results: directory where all the generated checks and results will be stored. There, one folder per each run are going to be placed.cartocell_1: run 1 experiment folder.aug: image augmentation samples.charts:cartocell_1_jaccard_index.png: IoU (jaccard_index) over epochs plot (when training is done).cartocell_1_loss.png: loss over epochs plot (when training is done).model_plot_cartocell_1.png: plot of the model.
per_image:.tif files: reconstructed images from patches.
per_image_instances:.tif files: same asper_imagebut with the instances.
per_image_post_processing:.tif files: same asper_image_instancesbut applied Voronoi, which has been the unique post-proccessing applied here.
watershed:seed_map.tif: initial seeds created before growing.foreground.tif: foreground mask area that delimits the grown of the seeds.watershed.tif: result of watershed.